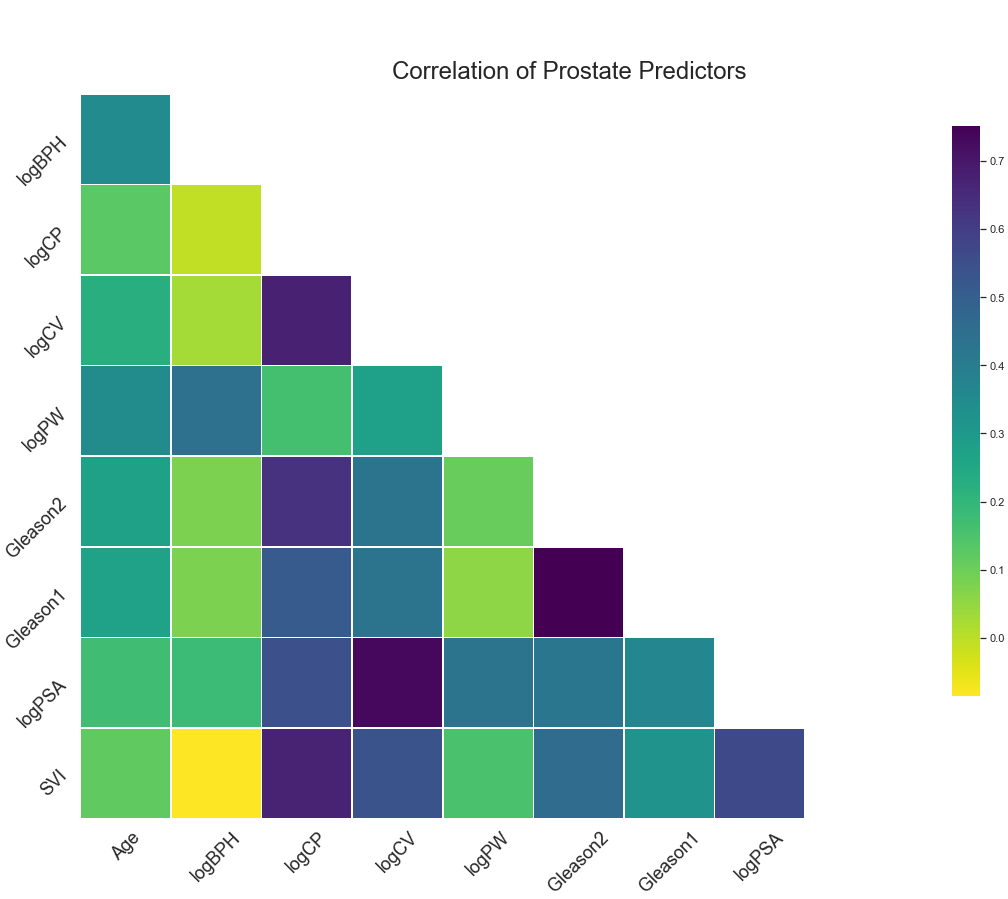
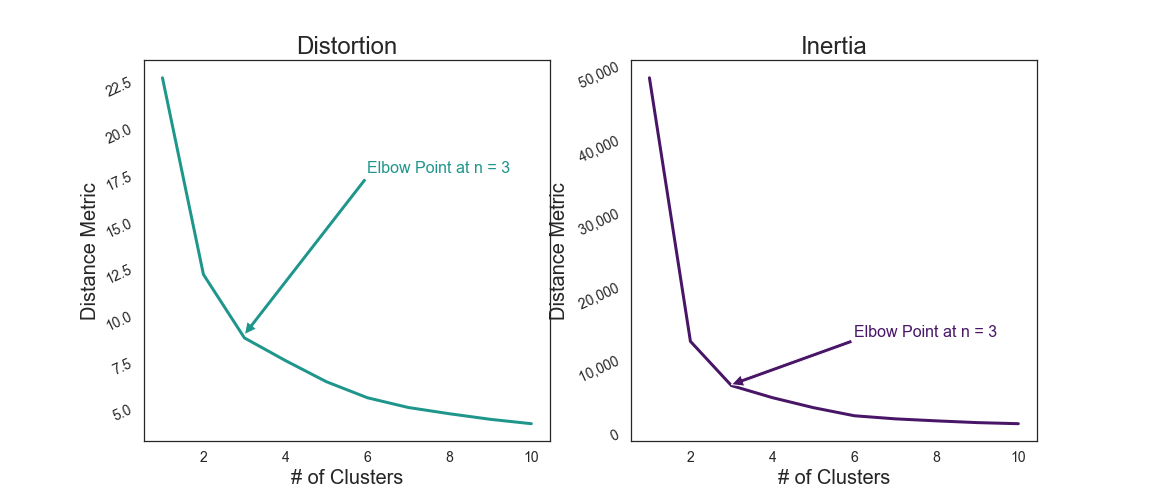
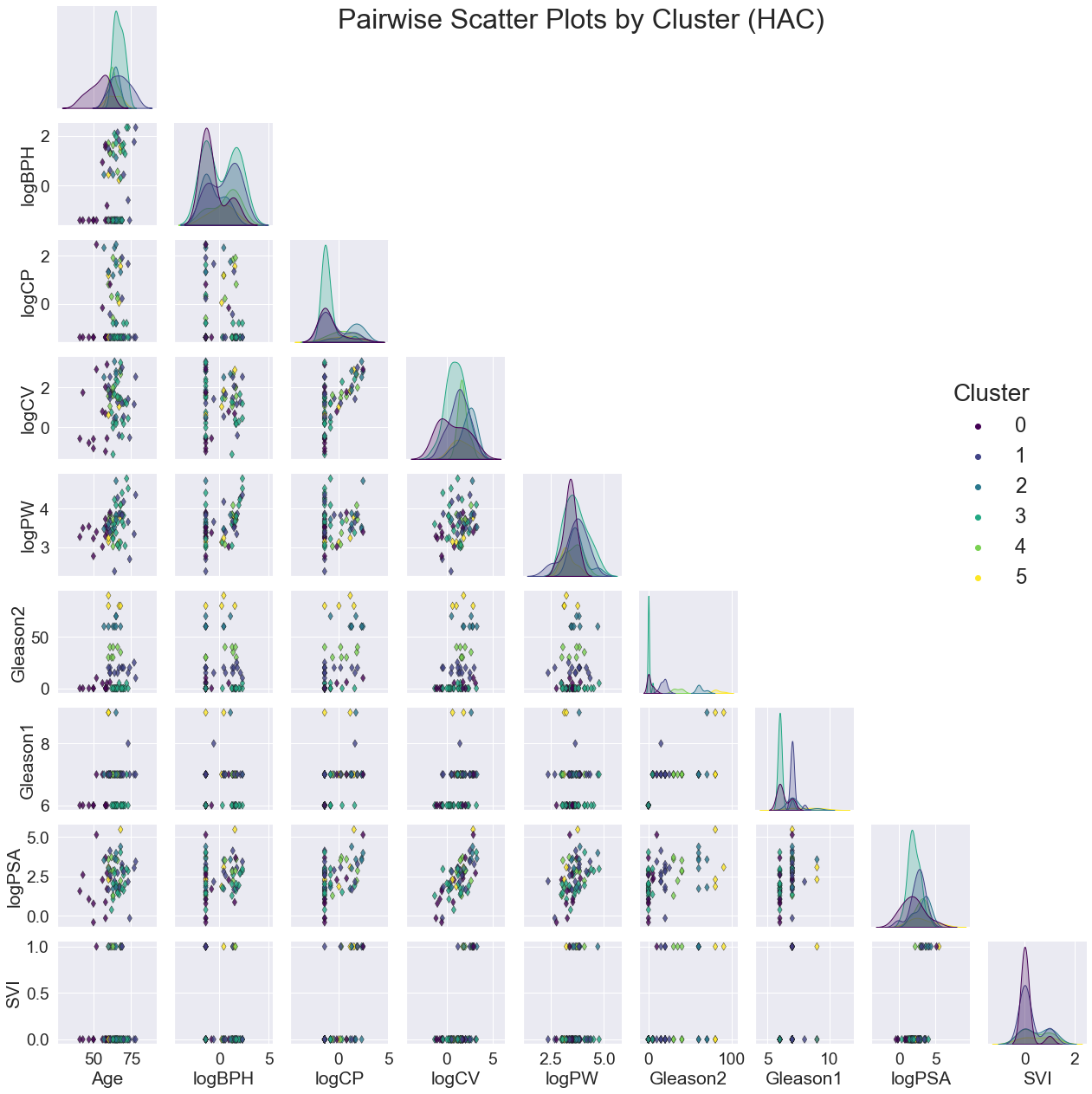
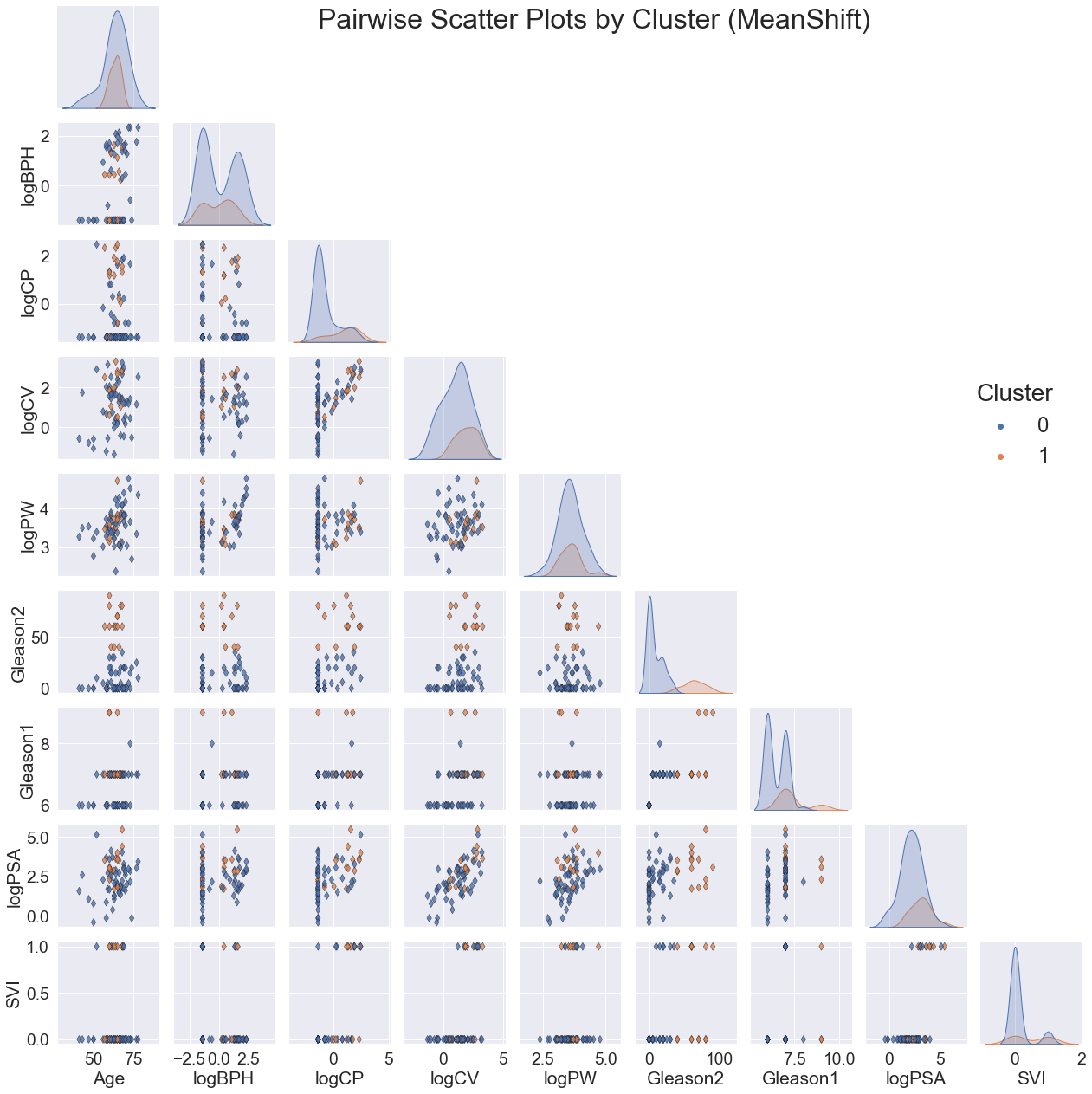
VI. Key Findings¶
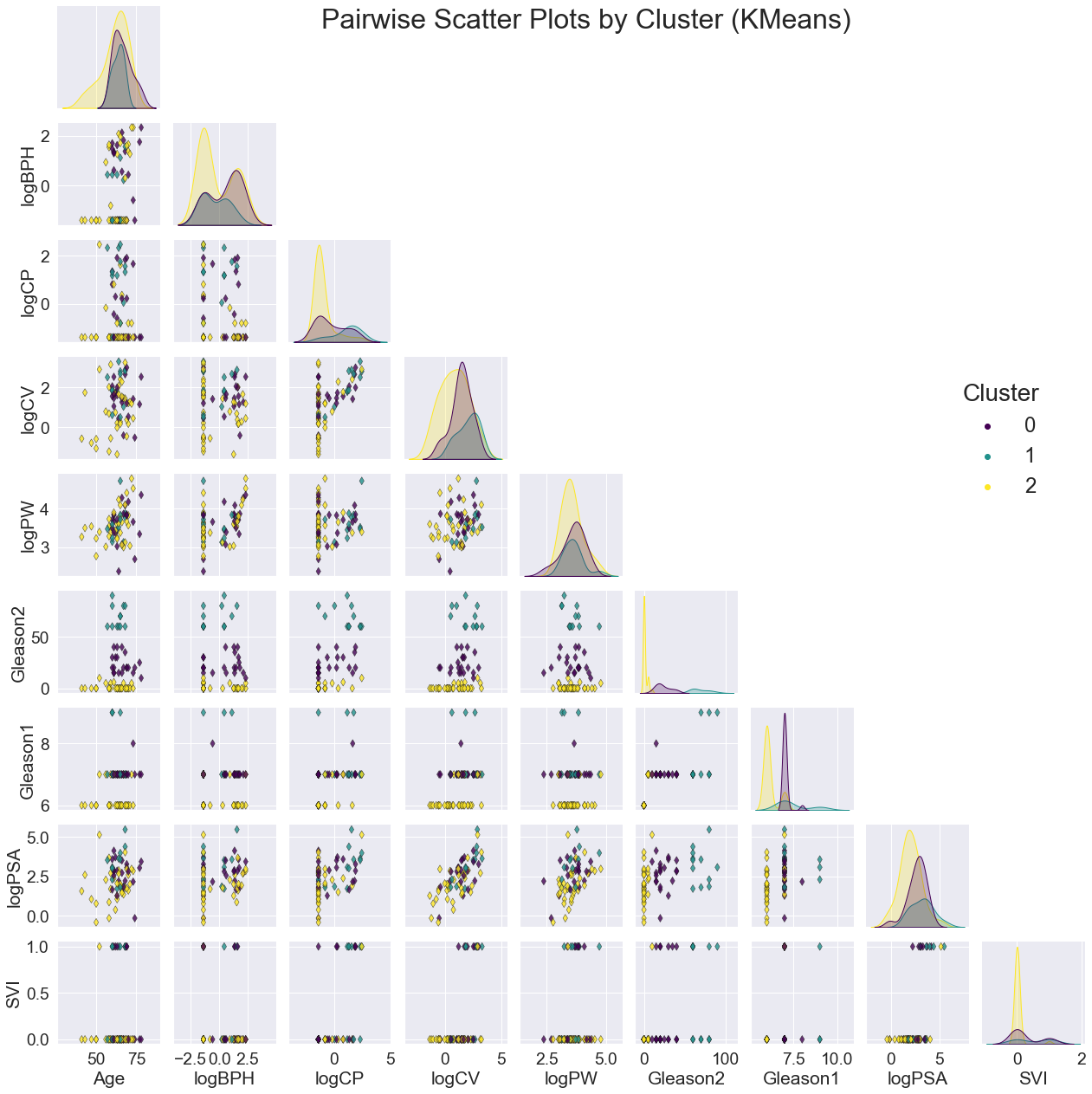
For all three models, the clusters seem to be most noticeably correlated with Gleason2. This is clear not only from the scatter plots of variable pairs, but the distinct separations in the distribution plot of Gleason2 along the diagonal.
Although there were a few peaks for the other variables, the overall distributions were generally highly overlapping. The scatter plots also had no emerging patterns.
VII. Final Model Selection¶
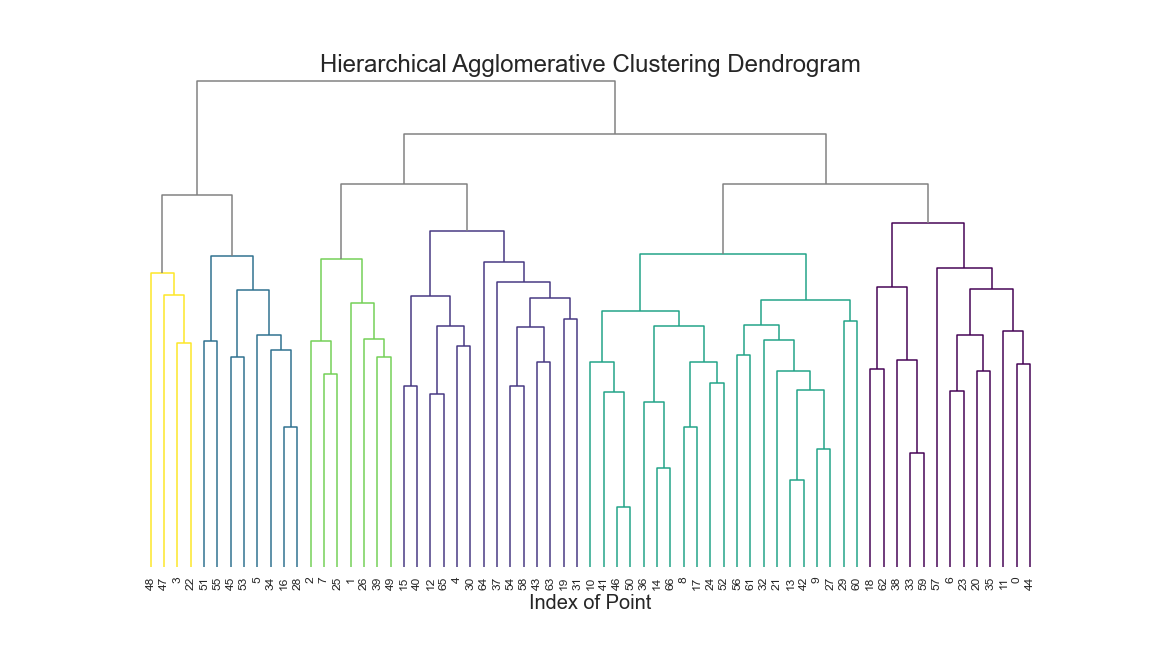
I choose Hierarchical Agglomerative Clustering as my final model. I chose it because I could set the cluster size from the dendogram and because it is recommended for uneven cluster sizes.
The validation pair-plots are shown below.
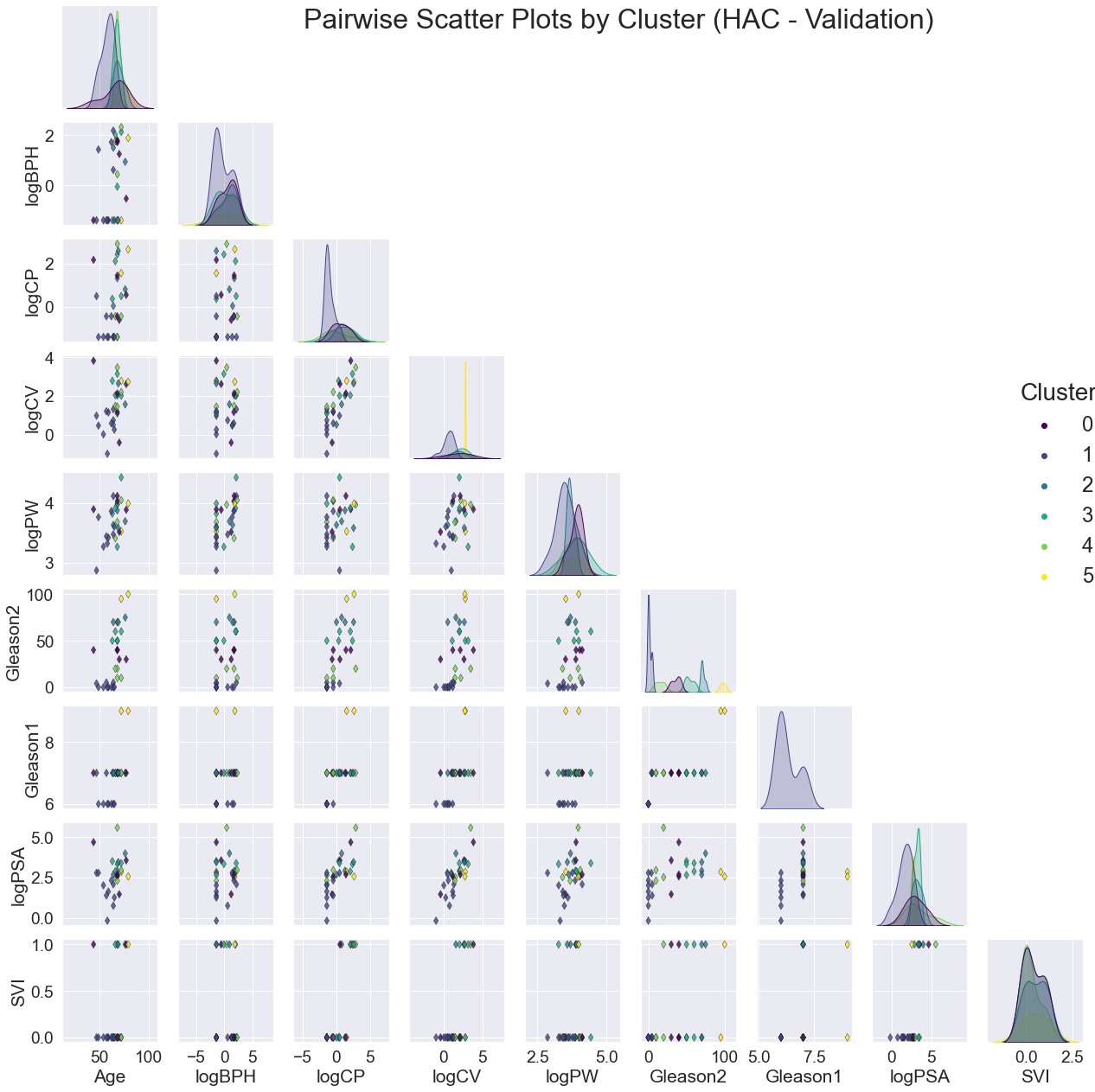
Again, the clusters are noticeably correlated with Gleason2. Perhaps Gleason2 is a good 'average' metric of the rest, which would explain more variance of the other variables among the clusters.
VIII. Conclusion, Next Steps, and Shortcomings¶
The dataset was only 97 observations and 9 variables. Only 67 observations were used in the clustering algorithms. More observations would have more diversity and perhaps more distinct clusters emerging.
Besides a few hours of reading some websites, I have very little subject knowledge of prostate health. I am certain there are many interactions, and perhaps other variables that I am unaware of which would be useful in clustering
Several variables were already log transformed. Would keeping their original values be more useful in clustering?
I did not fully test different affinities, linkages, or cluster numbers for the Hierarchical Agglomerative Clustering Algorithm. It is possible that some other combination of these parameters/hyperparameters would have more accurate clusters for diagnosis and treatment. A more in-depth look at the algorithm with subject knowledge would be very useful for better clustering.